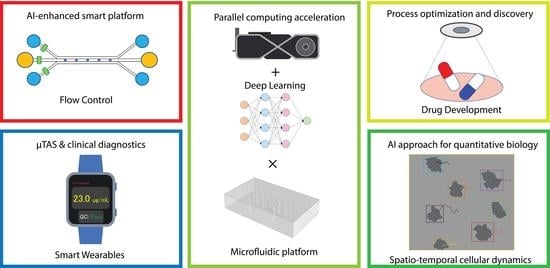
Microsystem Advances through Integration with Artificial Intelligence
Abstract
:1. Introduction
2. AI-Enhanced Smart Platform and Automation
2.1. Computer-Aided Microsystem Design and Optimization
Computational Fluid Dynamics (CFD) Modeling
2.2. Automation Control in Microsystems
2.2.1. Flow Control
2.2.2. Thermal Control

2.2.3. Particle Manipulation
2.2.4. Droplet Control, Detection, and Tracking
3. Process Optimization and Discovery
3.1. Synthetic Reaction Optimization
3.2. Nanoparticle Synthesis

3.3. Drug Development
4. Micro-Total Analysis System (TAS) and Clinical Diagnostics
4.1. Disease Diagnosis and Prognosis
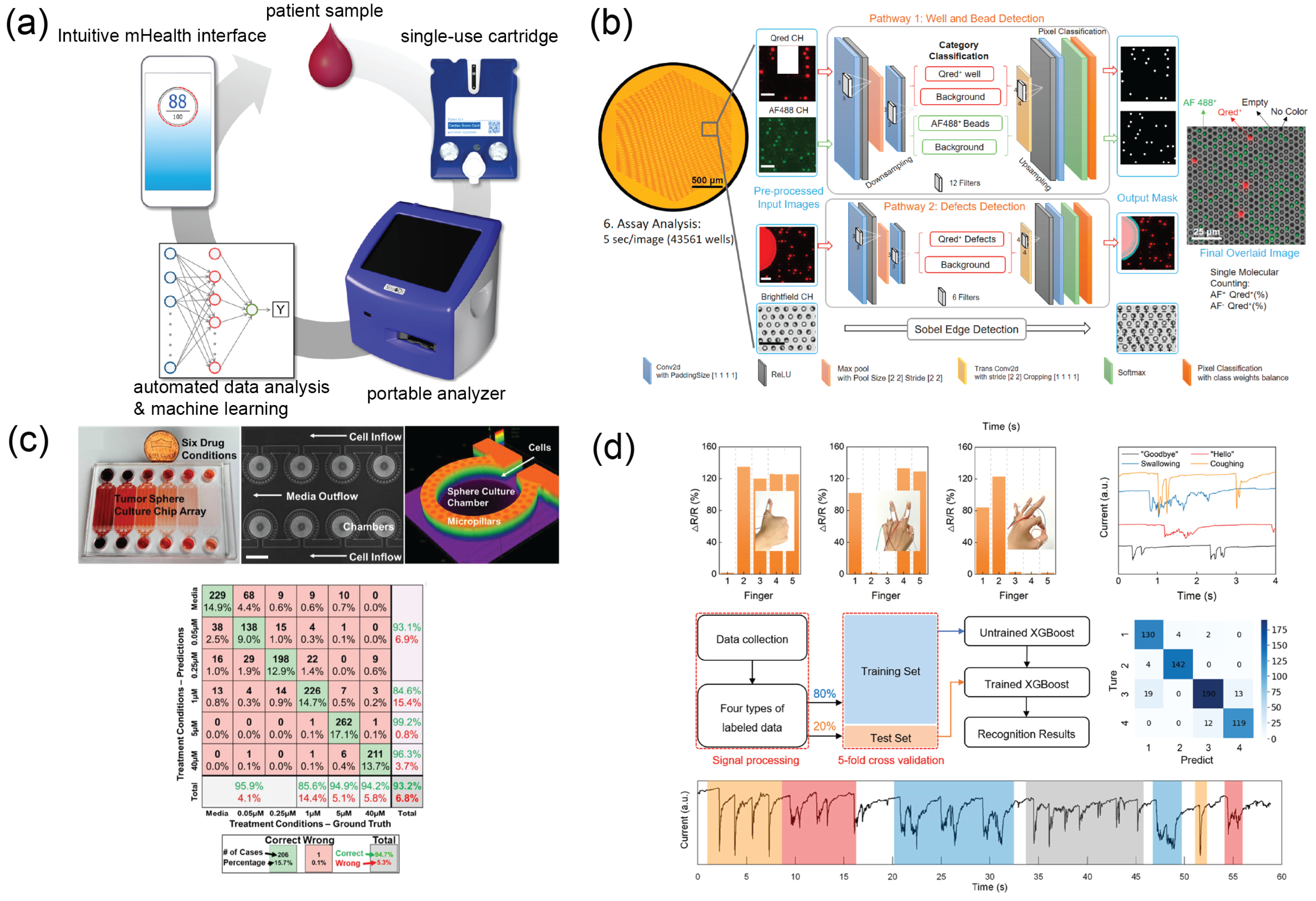
4.2. Drug Susceptibility Testing
4.3. Smart Wearables
5. AI Approach for Quantitative Biology
5.1. Cell Analysis
5.1.1. Cell Counting and Classification
5.1.2. Cell Sorting
5.1.3. Cell Phenotype Analysis
5.1.4. Spatiotemporal Cellular Dynamics Analysis

5.2. Personalized Medicine
5.2.1. Integration with Molecular Bioinformatics
5.2.2. Organ-on-Chips as Human Mimetic Models
5.2.3. Personalized Drug Development

6. Discussion and Outlook
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| AIV | Artificial Intelligence Velocimetry |
| ANN | Artificial Neural Network |
| AST | Antimicrobial Susceptibility Testing |
| CC-BY | Creative Commons Attribution 4.0 License |
| CFD | Computational Fluid Dynamics |
| CNN | Convolutional Neural Network |
| CTC | Circulating Tumor Cells |
| DM | Diabetes Mellitus |
| ELISA | Enzyme-Linked ImmunoSorbent Assay |
| FPS | Frames Per Second |
| GPU | Graphics Processing Unit |
| ICT | Information and Communication Technology |
| iPSC | Induced Pluripotent Stem Cell |
| IR | Infrared |
| LAMP | Loop-mediated isothermal AMPlification |
| MEMS | Microelectromechanical systems |
| MRD | Minimum Residual Disease |
| TAS | Micro-Total Analysis System |
| OoC | Organ-on-Chip |
| POCT | Point-Of-Care Testing |
| QPI | Quantitative Phase Imaging |
| RBC | Red Blood Cell |
| RNN | Recurrent Neural Network |
| SNR | Signal-to-Noise Ratio |
| SVM | Support Vector Machine |
| TDM | Therapeutic Drug Monitoring |
| WBC | White Blood Cell |
| YOLO | You Only Look Once |
References
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Stone, H.A. Introduction to Fluid Dynamics for Microfluidic Flows. In CMOS Biotechnology; Lee, H., Westervelt, R.M., Ham, D., Eds.; Series on Integrated Circuits and Systems; Springer: Boston, MA, USA, 2007; pp. 5–30. [Google Scholar]
- deMello, A.J. Control and detection of chemical reactions in microfluidic systems. Nature 2006, 442, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Velve-Casquillas, G.; Le Berre, M.; Piel, M.; Tran, P.T. Microfluidic tools for cell biological research. Nano Today 2010, 5, 28–47. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, X. Why microfluidics? Merits and trends in chemical synthesis. Lab Chip 2017, 17, 3960–3978. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.B.; Hanson, R.L.; Almughamsi, H.M.; Pang, C.; Fish, T.R.; Woolley, A.T. Microfluidics: Innovations in Materials and Their Fabrication and Functionalization. Anal. Chem. 2020, 92, 150–168. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Uhrig, R. Introduction to artificial neural networks. In Proceedings of the IECON ’95-21st Annual Conference on IEEE Industrial Electronics, Orlando, FL, USA, 6–10 November 1995; Volume 1, pp. 33–37. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural networks and their applications. Rev. Sci. Instrum. 1994, 65, 1803–1832. [Google Scholar] [CrossRef]
- Ben-Nun, T.; Hoefler, T. Demystifying Parallel and Distributed Deep Learning: An In-depth Concurrency Analysis. ACM Comput. Surv. 2019, 52, 65:1–65:43. [Google Scholar] [CrossRef]
- Rao, Q.; Frtunikj, J. Deep learning for self-driving cars: Chances and challenges. In Proceedings of the 1st International Workshop on Software Engineering for AI in Autonomous Systems (SEFAIS’18), Gothenburg, Sweden, 28 May 2018; Association for Computing Machinery: New York, NY, USA, 2018; pp. 35–38. [Google Scholar] [CrossRef]
- Johnson, J. Artificial intelligence & future warfare: Implications for international security. Def. Secur. Anal. 2019, 35, 147–169. [Google Scholar] [CrossRef]
- Yu, T.C.; Chou, W.C.; Yeh, C.Y.; Yang, C.K.; Huang, S.C.; Tien, F.M.; Yao, C.Y.; Cheng, C.L.; Chuang, M.K.; Tien, H.F.; et al. Automatic Bone Marrow Cell Identification and Classification By Deep Neural Network. Blood 2019, 134, 2084. [Google Scholar] [CrossRef]
- Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 2021, 5, 555–570. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Chen, C.C.; Yu, W.H.; Chen, S.H.; Chang, Y.C.; Hsu, T.I.; Hsiao, M.; Yeh, C.Y.; Chen, C.Y. An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning. Nat. Commun. 2021, 12, 1193. [Google Scholar] [CrossRef] [PubMed]
- Podder, S.; Bhattacharjee, S.; Roy, A.; Podder, S.; Bhattacharjee, S.; Roy, A. An efficient method of detection of COVID-19 using Mask R-CNN on chest X-Ray images. AIMS Biophys. 2021, 8, 281–290. [Google Scholar] [CrossRef]
- Galan, E.A.; Zhao, H.; Wang, X.; Dai, Q.; Huck, W.T.S.; Ma, S. Intelligent Microfluidics: The Convergence of Machine Learning and Microfluidics in Materials Science and Biomedicine. Matter 2020, 3, 1893–1922. [Google Scholar] [CrossRef]
- Bai, Y.; Gao, M.; Wen, L.; He, C.; Chen, Y.; Liu, C.; Fu, X.; Huang, S. Applications of Microfluidics in Quantitative Biology. Biotechnol. J. 2018, 13, 1700170. [Google Scholar] [CrossRef]
- McIntyre, D.; Lashkaripour, A.; Fordyce, P.; Densmore, D. Machine learning for microfluidic design and control. Lab Chip 2022, 22, 2925–2937. [Google Scholar] [CrossRef]
- Renner, G.; Ekárt, A. Genetic algorithms in computer aided design. Comput.-Aided Des. 2003, 35, 709–726. [Google Scholar] [CrossRef]
- Oh, K.W.; Lee, K.; Ahn, B.; Furlani, E.P. Design of pressure-driven microfluidic networks using electric circuit analogy. Lab Chip 2012, 12, 515–545. [Google Scholar] [CrossRef]
- Xu, H.; Liu, R.; Choudhary, A.; Chen, W. A Machine Learning-Based Design Representation Method for Designing Heterogeneous Microstructures. J. Mech. Des. 2015, 137. [Google Scholar] [CrossRef]
- Bhargava, K.C.; Thompson, B.; Iqbal, D.; Malmstadt, N. Predicting the behavior of microfluidic circuits made from discrete elements. Sci. Rep. 2015, 5, 15609. [Google Scholar] [CrossRef] [PubMed]
- Tsur, E.E. Computer-Aided Design of Microfluidic Circuits. Annu. Rev. Biomed. Eng. 2020, 22, 285–307. [Google Scholar] [CrossRef] [PubMed]
- Lore, K.G.; Stoecklein, D.; Davies, M.; Ganapathysubramanian, B.; Sarkar, S. Hierarchical Feature Extraction for Efficient Design of Microfluidic Flow Patterns. In Proceedings of the 1st International Workshop on Feature Extraction: Modern Questions and Challenges at NIPS2015, Montreal, QC, Canada, 11 December 2015; PMLR: Cambridge, MA, USA, 2015; pp. 213–225. [Google Scholar]
- Stoecklein, D.; Lore, K.G.; Davies, M.; Sarkar, S.; Ganapathysubramanian, B. Deep Learning for Flow Sculpting: Insights into Efficient Learning using Scientific Simulation Data. Sci. Rep. 2017, 7, 46368. [Google Scholar] [CrossRef]
- Lee, X.Y.; Balu, A.; Stoecklein, D.; Ganapathysubramanian, B.; Sarkar, S. Flow Shape Design for Microfluidic Devices Using Deep Reinforcement Learning. arXiv 2018. [Google Scholar] [CrossRef]
- Granados-Ortiz, F.J.; Ortega-Casanova, J. Machine learning-aided design optimization of a mechanical micromixer. Phys. Fluids 2021, 33, 063604. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, N.; Chen, J.; Su, G.; Yao, H.; Ho, T.Y.; Sun, L. Predicting the fluid behavior of random microfluidic mixers using convolutional neural networks. Lab Chip 2021, 21, 296–309. [Google Scholar] [CrossRef]
- Maionchi, D.d.O.; Ainstein, L.; dos Santos, F.P.; de Souza Júnior, M.B. Computational fluid dynamics and machine learning as tools for optimization of micromixers geometry. Int. J. Heat Mass Transf. 2022, 194, 123110. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, Z.; Wang, J. Machine-Learning-Enabled Design and Manipulation of a Microfluidic Concentration Gradient Generator. Micromachines 2022, 13, 1810. [Google Scholar] [CrossRef] [PubMed]
- Balabanov, A.V.; Kasimov, A.M.; Popov, A.I.; Fateev, V.Y. MNM-Modelling and Creating Designs of Discrete Microfluidics. In Proceedings of the 2021 14th International Conference Management of Large-Scale System Development (MLSD), Moscow, Russian, 27–29 September 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Lashkaripour, A.; Rodriguez, C.; Mehdipour, N.; Mardian, R.; McIntyre, D.; Ortiz, L.; Campbell, J.; Densmore, D. Machine learning enables design automation of microfluidic flow-focusing droplet generation. Nat. Commun. 2021, 12, 25. [Google Scholar] [CrossRef]
- Shahab, M.; Rengaswamy, R. Reinforcement-Learning designs droplet microfluidic networks. Comput. Chem. Eng. 2022, 161, 107787. [Google Scholar] [CrossRef]
- Haward, S.J.; Oliveira, M.S.N.; Alves, M.A.; McKinley, G.H. Optimized Cross-Slot Flow Geometry for Microfluidic Extensional Rheometry. Phys. Rev. Lett. 2012, 109, 128301. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liang, K.; Zhang, N.; Yao, H.; Ho, T.Y.; Sun, L. Automated calibration of 3D-printed microfluidic devices based on computer vision. Biomicrofluidics 2021, 15, 024102. [Google Scholar] [CrossRef] [PubMed]
- Shchanikov, S.; Zuev, A.; Bordanov, I.; Danilin, S.; Lukoyanov, V.; Korolev, D.; Belov, A.; Pigareva, Y.; Gladkov, A.; Pimashkin, A.; et al. Designing a bidirectional, adaptive neural interface incorporating machine learning capabilities and memristor-enhanced hardware. Chaos Solitons Fractals 2021, 142, 110504. [Google Scholar] [CrossRef]
- Bachratý, H.; Bachratá, K.; Chovanec, M.; Jančigová, I.; Smiešková, M.; Kovalčíková, K. Applications of machine learning for simulations of red blood cells in microfluidic devices. BMC Bioinform. 2020, 21, 90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Liang, K.; Liu, Z.; Sun, T.; Wang, J. ANN-Based Instantaneous Simulation of Particle Trajectories in Microfluidics. Micromachines 2022, 13, 2100. [Google Scholar] [CrossRef]
- Ahmed, F.; Shimizu, M.; Wang, J.; Sakai, K.; Kiwa, T. Optimization of Microchannels and Application of Basic Activation Functions of Deep Neural Network for Accuracy Analysis of Microfluidic Parameter Data. Micromachines 2022, 13, 1352. [Google Scholar] [CrossRef] [PubMed]
- Kutz, J.N. Deep learning in fluid dynamics. J. Fluid Mech. 2017, 814, 1–4. [Google Scholar] [CrossRef]
- Brunton, S.L.; Noack, B.R.; Koumoutsakos, P. Machine Learning for Fluid Mechanics. Annu. Rev. Fluid Mech. 2020, 52, 477–508. [Google Scholar] [CrossRef]
- Kochkov, D.; Smith, J.A.; Alieva, A.; Wang, Q.; Brenner, M.P.; Hoyer, S. Machine learning–accelerated computational fluid dynamics. Proc. Natl. Acad. Sci. USA 2021, 118, e2101784118. [Google Scholar] [CrossRef]
- Cai, S.; Li, H.; Zheng, F.; Kong, F.; Dao, M.; Karniadakis, G.E.; Suresh, S. Artificial intelligence velocimetry and microaneurysm-on-a-chip for three-dimensional analysis of blood flow in physiology and disease. Proc. Natl. Acad. Sci. USA 2021, 118, e2100697118. [Google Scholar] [CrossRef]
- Zeng, X.; Xue, C.D.; Chen, K.J.; Li, Y.J.; Qin, K.R. Deep-learning-assisted extraction of height-averaged velocity from scalar signal transport in a shallow microfluidic channel. Microfluid. Nanofluidics 2022, 26, 36. [Google Scholar] [CrossRef]
- Chen, Q.; Deng, J.; Luo, G. Micromixing Performance and Residence Time Distribution in a Miniaturized Magnetic Reactor: Experimental Investigation and Machine Learning Modeling. Ind. Eng. Chem. Res. 2023, 62, 3577–3591. [Google Scholar] [CrossRef]
- Sharma, H.; Das, G.; Samanta, A.N. ANN–based prediction of two-phase gas– liquid flow patterns in a circular conduit. AIChE J. 2006, 52, 3018–3028. [Google Scholar] [CrossRef]
- Giri Nandagopal, M.S.; Selvaraju, N. Prediction of Liquid–Liquid Flow Patterns in a Y-Junction Circular Microchannel Using Advanced Neural Network Techniques. Ind. Eng. Chem. Res. 2016, 55, 11346–11362. [Google Scholar] [CrossRef]
- Nandagopal, M.S.G.; Abraham, E.; Selvaraju, N. Advanced neural network prediction and system identification of liquid-liquid flow patterns in circular microchannels with varying angle of confluence. Chem. Eng. J. 2017, 309, 850–865. [Google Scholar] [CrossRef]
- Desir, P.; Chen, T.Y.; Bracconi, M.; Saha, B.; Maestri, M.; Vlachos, D.G. Experiments and computations of microfluidic liquid–liquid flow patterns. React. Chem. Eng. 2019, 5, 39–50. [Google Scholar] [CrossRef]
- Abbasi Moud, A. Recent advances in utility of artificial intelligence towards multiscale colloidal based materials design. Colloid Interface Sci. Commun. 2022, 47, 100595. [Google Scholar] [CrossRef]
- Iverson, B.D.; Garimella, S.V. Recent advances in microscale pumping technologies: A review and evaluation. Microfluid. Nanofluidics 2008, 5, 145–174. [Google Scholar] [CrossRef]
- Dressler, O.J.; Howes, P.D.; Choo, J.; deMello, A.J. Reinforcement Learning for Dynamic Microfluidic Control. ACS Omega 2018, 3, 10084–10091. [Google Scholar] [CrossRef]
- Au, A.K.; Lai, H.; Utela, B.R.; Folch, A. Microvalves and Micropumps for BioMEMS. Micromachines 2011, 2, 179–220. [Google Scholar] [CrossRef]
- Abe, T.; Oh-hara, S.; Ukita, Y. Adoption of reinforcement learning for the intelligent control of a microfluidic peristaltic pump. Biomicrofluidics 2021, 15, 034101. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Oh-hara, S.; Ukita, Y. Integration of reinforcement learning to realize functional variability of microfluidic systems. Biomicrofluidics 2022, 16, 024106. [Google Scholar] [CrossRef]
- Shayan, M.; Bhattacharjee, S.; Song, Y.A.; Chakrabarty, K.; Karri, R. Toward Secure Microfluidic Fully Programmable Valve Array Biochips. IEEE Trans. Very Large Scale Integr. (VLSI) Syst. 2019, 27, 2755–2766. [Google Scholar] [CrossRef]
- Hajmohammadi, M.R.; Alipour, P.; Parsa, H. Microfluidic effects on the heat transfer enhancement and optimal design of microchannels heat sinks. Int. J. Heat Mass Transf. 2018, 126, 808–815. [Google Scholar] [CrossRef]
- Miralles, V.; Huerre, A.; Malloggi, F.; Jullien, M.C. A Review of Heating and Temperature Control in Microfluidic Systems: Techniques and Applications. Diagnostics 2013, 3, 33–67. [Google Scholar] [CrossRef]
- Rizkin, B.A.; Popovich, K.; Hartman, R.L. Artificial Neural Network control of thermoelectrically-cooled microfluidics using computer vision based on IR thermography. Comput. Chem. Eng. 2019, 121, 584–593. [Google Scholar] [CrossRef]
- Quinn, D.; Thalheim, T.; Cichos, F. Microfluidics with feedback control and machine learning (Conference Presentation). In Proceedings of the Emerging Topics in Artificial Intelligence (ETAI) 2022, San Diego, CA, USA, 21–25 August 2022; SPIE: Bellingham, WA, USA, 2022; Volume PC12204, p. PC122040S. [Google Scholar] [CrossRef]
- Lewis, C.; Erikson, J.W.; Sanchez, D.A.; McClure, C.E.; Nordin, G.P.; Munro, T.R.; Colton, J.S. Use of Machine Learning with Temporal Photoluminescence Signals from CdTe Quantum Dots for Temperature Measurement in Microfluidic Devices. ACS Appl. Nano Mater. 2020, 3, 4045–4053. [Google Scholar] [CrossRef] [PubMed]
- Raymond, S.J.; Collins, D.J.; O’Rorke, R.; Tayebi, M.; Ai, Y.; Williams, J. A deep learning approach for designed diffraction-based acoustic patterning in microchannels. Sci. Rep. 2020, 10, 8745. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Duan, F.; Zhou, A.; Kanitthamniyom, P.; Luo, S.; Hu, X.; Jiang, X.; Vasoo, S.; Zhang, X.; Zhang, Y. Image-based real-time feedback control of magnetic digital microfluidics by artificial intelligence-empowered rapid object detector for automated in vitro diagnostics. Bioeng. Transl. Med. 2022, e10428. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Onck, P.; den Toonder, J. A concise review of microfluidic particle manipulation methods. Microfluid. Nanofluidics 2020, 24, 24. [Google Scholar] [CrossRef]
- Fang, W.Z.; Xiong, T.; Pak, O.S.; Zhu, L. Data-Driven Intelligent Manipulation of Particles in Microfluidics. Adv. Sci. 2022, 10, 2205382. [Google Scholar] [CrossRef]
- Bazaz, S.R.; Mashhadian, A.; Ehsani, A.; Saha, S.C.; Krüger, T.; Warkiani, M.E. Computational inertial microfluidics: A review. Lab Chip 2020, 20, 1023–1048. [Google Scholar] [CrossRef] [PubMed]
- Del Giudice, F. A Review of Microfluidic Devices for Rheological Characterisation. Micromachines 2022, 13, 167. [Google Scholar] [CrossRef]
- Su, J.; Chen, X.; Zhu, Y.; Hu, G. Machine learning assisted fast prediction of inertial lift in microchannels. Lab Chip 2021, 21, 2544–2556. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, E.; Dezhkam, R.; Shamloo, A.; Mashhadian, A. microAI: A machine learning tool for fast calculation of lift coefficients in microchannels. arXiv 2022. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, A.; Chen, S.; Lum, G.Z.; Zhang, X. A perspective on magnetic microfluidics: Towards an intelligent future. Biomicrofluidics 2022, 16, 011301. [Google Scholar] [CrossRef]
- Koh, J.B.Y.; Shen, X.; Marcos. Supervised Learning to Predict Sperm Sorting by Magnetophoresis. Magnetochemistry 2018, 4, 31. [Google Scholar] [CrossRef]
- Ciriza, D.B.; Magazzù, A.; Callegari, A.; Iatì, M.A.; Volpe, G.; Maragò, O.M. Machine learning to enhance the calculation of optical forces in the geometrical optics approximation. In Proceedings of the Biophotonics Congress 2021, Washington, DC, USA, 12–16 April 2021; Optica Publishing Group: Washington, DC, USA, 2021; p. AF2D.3. [Google Scholar] [CrossRef]
- Zhao, J.; Hou, H.; Huang, Q.Y.; Zhong, X.G.; Zheng, P.S. Design of Optical Tweezers Manipulation Control System Based on Novel Self-Organizing Fuzzy Cerebellar Model Neural Network. Appl. Sci. 2022, 12, 9655. [Google Scholar] [CrossRef]
- Harshbarger, C.L.; Gerlt, M.S.; Ghadamian, J.A.; Bernardoni, D.C.; Snedeker, J.G.; Dual, J. Optical feedback control loop for the precise and robust acoustic focusing of cells, micro- and nanoparticles. Lab Chip 2022, 22, 2810–2819. [Google Scholar] [CrossRef]
- Yiannacou, K.; Sariola, V. Controlled Manipulation and Active Sorting of Particles Inside Microfluidic Chips Using Bulk Acoustic Waves and Machine Learning. Langmuir 2021, 37, 4192–4199. [Google Scholar] [CrossRef]
- Yiannacou, K.; Sharma, V.; Sariola, V. Programmable Droplet Microfluidics Based on Machine Learning and Acoustic Manipulation. Langmuir 2022, 38, 11557–11564. [Google Scholar] [CrossRef]
- Teh, S.Y.; Lin, R.; Hung, L.H.; Lee, A.P. Droplet microfluidics. Lab Chip 2008, 8, 198–220. [Google Scholar] [CrossRef]
- Pouyanfar, N.; Harofte, S.Z.; Soltani, M.; Siavashy, S.; Asadian, E.; Ghorbani-Bidkorbeh, F.; Keçili, R.; Hussain, C.M. Artificial intelligence-based microfluidic platforms for the sensitive detection of environmental pollutants: Recent advances and prospects. Trends Environ. Anal. Chem. 2022, 34, e00160. [Google Scholar] [CrossRef]
- Mahdi, Y.; Daoud, K. Microdroplet size prediction in microfluidic systems via artificial neural network modeling for water-in-oil emulsion formulation. J. Dispers. Sci. Technol. 2017, 38, 1501–1508. [Google Scholar] [CrossRef]
- Crowson, M.G.; Moukheiber, D.; Arévalo, A.R.; Lam, B.D.; Mantena, S.; Rana, A.; Goss, D.; Bates, D.W.; Celi, L.A. A systematic review of federated learning applications for biomedical data. PLoS Digit. Health 2022, 1, e0000033. [Google Scholar] [CrossRef] [PubMed]
- Siemenn, A.E.; Shaulsky, E.; Beveridge, M.; Buonassisi, T.; Hashmi, S.M.; Drori, I. A Machine Learning and Computer Vision Approach to Rapidly Optimize Multiscale Droplet Generation. ACS Appl. Mater. Interfaces 2022, 14, 4668–4679. [Google Scholar] [CrossRef]
- Khor, J.W.; Jean, N.; Luxenberg, E.S.; Ermon, S.; Tang, S.K.Y. Using machine learning to discover shape descriptors for predicting emulsion stability in a microfluidic channel. Soft Matter 2019, 15, 1361–1372. [Google Scholar] [CrossRef]
- Chagot, L.; Quilodrán-Casas, C.; Kalli, M.; Kovalchuk, N.M.; Simmons, M.J.H.; Matar, O.K.; Arcucci, R.; Angeli, P. Surfactant-laden droplet size prediction in a flow-focusing microchannel: A data-driven approach. Lab Chip 2022, 22, 3848–3859. [Google Scholar] [CrossRef]
- Gardner, K.; Uddin, M.M.; Tran, L.; Pham, T.; Vanapalli, S.; Li, W. Deep learning detector for high precision monitoring of cell encapsulation statistics in microfluidic droplets. Lab Chip 2022, 22, 4067–4080. [Google Scholar] [CrossRef]
- Rutkowski, G.P.; Azizov, I.; Unmann, E.; Dudek, M.; Grimes, B.A. Microfluidic droplet detection via region-based and single-pass convolutional neural networks with comparison to conventional image analysis methodologies. Mach. Learn. Appl. 2022, 7, 100222. [Google Scholar] [CrossRef]
- Vaithiyanathan, M.; Safa, N.; Melvin, A.T. FluoroCellTrack: An algorithm for automated analysis of high-throughput droplet microfluidic data. PLoS ONE 2019, 14, e0215337. [Google Scholar] [CrossRef]
- Durve, M.; Tiribocchi, A.; Bonaccorso, F.; Montessori, A.; Lauricella, M.; Bogdan, M.; Guzowski, J.; Succi, S. DropTrack – automatic droplet tracking using deep learning for microfluidic applications. Phys. Fluids 2022, 34, 082003. [Google Scholar] [CrossRef]
- Zhuang, Y.; Cheng, S.; Kovalchuk, N.; Simmons, M.; Matar, O.K.; Guo, Y.K.; Arcucci, R. Ensemble latent assimilation with deep learning surrogate model: Application to drop interaction in a microfluidics device. Lab Chip 2022, 22, 3187–3202. [Google Scholar] [CrossRef]
- Hadikhani, P.; Borhani, N.H.; Hashemi, S.M.; Psaltis, D. Learning from droplet flows in microfluidic channels using deep neural networks. Sci. Rep. 2019, 9, 8114. [Google Scholar] [CrossRef]
- Arjun, A.; Ajith, R.R.; Kumar Ranjith, S. Mixing characterization of binary-coalesced droplets in microchannels using deep neural network. Biomicrofluidics 2020, 14, 034111. [Google Scholar] [CrossRef]
- Roy, P.; House, M.L.; Dutcher, C.S. A Microfluidic Device for Automated High Throughput Detection of Ice Nucleation of Snomax®. Micromachines 2021, 12, 296. [Google Scholar] [CrossRef]
- Liang, T.C.; Zhong, Z.; Bigdeli, Y.; Ho, T.Y.; Chakrabarty, K.; Fair, R. Adaptive Droplet Routing in Digital Microfluidic Biochips Using Deep Reinforcement Learning. In Proceedings of the 37th International Conference on Machine Learning, Virtual, 13–18 July 2020; PMLR: Cambridge, MA, USA, 2020; pp. 6050–6060. [Google Scholar]
- Jiang, C.; Yang, R.Q.; Yuan, B. An evolutionary algorithm with indirect representation for droplet routing in digital microfluidic biochips. Eng. Appl. Artif. Intell. 2022, 115, 105305. [Google Scholar] [CrossRef]
- Chu, A.; Nguyen, D.; Talathi, S.S.; Wilson, A.C.; Ye, C.; Smith, W.L.; Kaplan, A.D.; Duoss, E.B.; Stolaroff, J.K.; Giera, B. Automated detection and sorting of microencapsulation via machine learning. Lab Chip 2019, 19, 1808–1817. [Google Scholar] [CrossRef] [PubMed]
- Anagnostidis, V.; Sherlock, B.; Metz, J.; Mair, P.; Hollfelder, F.; Gielen, F. Deep learning guided image-based droplet sorting for on-demand selection and analysis of single cells and 3D cell cultures. Lab Chip 2020, 20, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Howell, L.; Anagnostidis, V.; Gielen, F. Multi-Object Detector YOLOv4-Tiny Enables High-Throughput Combinatorial and Spatially-Resolved Sorting of Cells in Microdroplets. Adv. Mater. Technol. 2022, 7, 2101053. [Google Scholar] [CrossRef]
- Liu, L.; Bi, M.; Wang, Y.; Liu, J.; Jiang, X.; Xu, Z.; Zhang, X. Artificial intelligence-powered microfluidics for nanomedicine and materials synthesis. Nanoscale 2021, 13, 19352–19366. [Google Scholar] [CrossRef]
- Reizman, B.J.; Jensen, K.F. Feedback in Flow for Accelerated Reaction Development. Acc. Chem. Res. 2016, 49, 1786–1796. [Google Scholar] [CrossRef]
- Granda, J.M.; Donina, L.; Dragone, V.; Long, D.L.; Cronin, L. Controlling an organic synthesis robot with machine learning to search for new reactivity. Nature 2018, 559, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Riordon, J.; Wu, T.C.; Edwards, H.; Wheeler, A.R.; Pardee, K.; Aspuru-Guzik, A.; Sinton, D. When robotics met fluidics. Lab Chip 2020, 20, 709–716. [Google Scholar] [CrossRef]
- McMullen, J.P.; Jensen, K.F. An Automated Microfluidic System for Online Optimization in Chemical Synthesis. Org. Process Res. Dev. 2010, 14, 1169–1176. [Google Scholar] [CrossRef]
- McMullen, J.P.; Stone, M.T.; Buchwald, S.L.; Jensen, K.F. An Integrated Microreactor System for Self-Optimization of a Heck Reaction: From Micro- to Mesoscale Flow Systems. Angew. Chem. Int. Ed. 2010, 49, 7076–7080. [Google Scholar] [CrossRef]
- Rizkin, B.A.; Shkolnik, A.S.; Ferraro, N.J.; Hartman, R.L. Combining automated microfluidic experimentation with machine learning for efficient polymerization design. Nat. Mach. Intell. 2020, 2, 200–209. [Google Scholar] [CrossRef]
- Chen, X.; Lv, H. Intelligent control of nanoparticle synthesis on microfluidic chips with machine learning. NPG Asia Mater. 2022, 14, 1–20. [Google Scholar] [CrossRef]
- Krishnadasan, S.; Brown, R.J.C.; deMello, A.J.; deMello, J.C. Intelligent routes to the controlled synthesis of nanoparticles. Lab Chip 2007, 7, 1434–1441. [Google Scholar] [CrossRef] [PubMed]
- Orimoto, Y.; Watanabe, K.; Yamashita, K.; Uehara, M.; Nakamura, H.; Furuya, T.; Maeda, H. Application of Artificial Neural Networks to Rapid Data Analysis in Combinatorial Nanoparticle Syntheses. J. Phys. Chem. C 2012, 116, 17885–17896. [Google Scholar] [CrossRef]
- Ahrberg, C.D.; Wook Choi, J.; Geun Chung, B. Automated droplet reactor for the synthesis of iron oxide/gold core-shell nanoparticles. Sci. Rep. 2020, 10, 1737. [Google Scholar] [CrossRef]
- Mekki-Berrada, F.; Ren, Z.; Huang, T.; Wong, W.K.; Zheng, F.; Xie, J.; Tian, I.P.S.; Jayavelu, S.; Mahfoud, Z.; Bash, D.; et al. Two-step machine learning enables optimized nanoparticle synthesis. Npj Comput. Mater. 2021, 7, 1–10. [Google Scholar] [CrossRef]
- Tao, H.; Wu, T.; Kheiri, S.; Aldeghi, M.; Aspuru-Guzik, A.; Kumacheva, E. Self-Driving Platform for Metal Nanoparticle Synthesis: Combining Microfluidics and Machine Learning. Adv. Funct. Mater. 2021, 31, 2106725. [Google Scholar] [CrossRef]
- Volk, A.A.; Epps, R.W.; Abolhasani, M. Accelerated Development of Colloidal Nanomaterials Enabled by Modular Microfluidic Reactors: Toward Autonomous Robotic Experimentation. Adv. Mater. 2021, 33, 2004495. [Google Scholar] [CrossRef]
- Wahl, C.B.; Aykol, M.; Swisher, J.H.; Montoya, J.H.; Suram, S.K.; Mirkin, C.A. Machine learning–accelerated design and synthesis of polyelemental heterostructures. Sci. Adv. 2021, 7, eabj5505. [Google Scholar] [CrossRef]
- Wang, S.; Shen, Z.; Shen, Z.; Dong, Y.; Li, Y.; Cao, Y.; Zhang, Y.; Guo, S.; Shuai, J.; Yang, Y.; et al. Machine-learning micropattern manufacturing. Nano Today 2021, 38, 101152. [Google Scholar] [CrossRef]
- Ali, H.S.M.; Blagden, N.; York, P.; Amani, A.; Brook, T. Artificial neural networks modelling the prednisolone nanoprecipitation in microfluidic reactors. Eur. J. Pharm. Sci. 2009, 37, 514–522. [Google Scholar] [CrossRef]
- Kirmani, A.R.; Luther, J.M.; Abolhasani, M.; Amassian, A. Colloidal Quantum Dot Photovoltaics: Current Progress and Path to Gigawatt Scale Enabled by Smart Manufacturing. ACS Energy Lett. 2020, 5, 3069–3100. [Google Scholar] [CrossRef]
- Chen, G.; Zhu, X.; Xing, C.; Wang, Y.; Xu, X.; Bao, J.; Huang, J.; Zhao, Y.; Wang, X.; Zhou, X.; et al. Machine Learning-Assisted Microfluidic Synthesis of Perovskite Quantum Dots. Adv. Photonics Res. 2022, 4, 2200230. [Google Scholar] [CrossRef]
- Rebollo, R.; Oyoun, F.; Corvis, Y.; El-Hammadi, M.M.; Saubamea, B.; Andrieux, K.; Mignet, N.; Alhareth, K. Microfluidic Manufacturing of Liposomes: Development and Optimization by Design of Experiment and Machine Learning. ACS Appl. Mater. Interfaces 2022, 14, 39736–39745. [Google Scholar] [CrossRef] [PubMed]
- Damiati, S.A.; Rossi, D.; Joensson, H.N.; Damiati, S. Artificial intelligence application for rapid fabrication of size-tunable PLGA microparticles in microfluidics. Sci. Rep. 2020, 10, 19517. [Google Scholar] [CrossRef] [PubMed]
- Damiati, S.A.; Damiati, S. Microfluidic Synthesis of Indomethacin-Loaded PLGA Microparticles Optimized by Machine Learning. Front. Mol. Biosci. 2021, 8, 595. [Google Scholar] [CrossRef] [PubMed]
- Grisoni, F.; Huisman, B.J.H.; Button, A.L.; Moret, M.; Atz, K.; Merk, D.; Schneider, G. Combining generative artificial intelligence and on-chip synthesis for de novo drug design. Sci. Adv. 2021, 7, eabg3338. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G. Automating drug discovery. Nat. Rev. Drug Discov. 2018, 17, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, L.; Zhang, H.; Shang, L.; Zhao, Y. Microfluidics for Drug Development: From Synthesis to Evaluation. Chem. Rev. 2021, 121, 7468–7529. [Google Scholar] [CrossRef]
- Popova, M.; Isayev, O.; Tropsha, A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018, 4, eaap7885. [Google Scholar] [CrossRef]
- Bannigan, P.; Bao, Z.; Hickman, R.J.; Aldeghi, M.; Häse, F.; Aspuru-Guzik, A.; Allen, C. Machine learning models to accelerate the design of polymeric long-acting injectables. Nat. Commun. 2023, 14, 35. [Google Scholar] [CrossRef]
- Mejía-Salazar, J.R.; Rodrigues Cruz, K.; Materón Vásques, E.M.; Novais de Oliveira, O., Jr. Microfluidic Point-of-Care Devices: New Trends and Future Prospects for eHealth Diagnostics. Sensors 2020, 20, 1951. [Google Scholar] [CrossRef]
- Zare Harofte, S.; Soltani, M.; Siavashy, S.; Raahemifar, K. Recent Advances of Utilizing Artificial Intelligence in Lab on a Chip for Diagnosis and Treatment. Small 2022, 18, 2203169. [Google Scholar] [CrossRef]
- Romao, V.C.; Martins, S.A.M.; Germano, J.; Cardoso, F.A.; Cardoso, S.; Freitas, P.P. Lab-on-Chip Devices: Gaining Ground Losing Size. ACS Nano 2017, 11, 10659–10664. [Google Scholar] [CrossRef]
- Dabbagh, S.R.; Rabbi, F.; Doğan, Z.; Yetisen, A.K.; Tasoglu, S. Machine learning-enabled multiplexed microfluidic sensors. Biomicrofluidics 2020, 14, 061506. [Google Scholar] [CrossRef]
- Lee, W.; Gonzalez, A.; Arguelles, P.; Guevara, R.; Gonzalez-Guerrero, M.J.; Gomez, F.A. Thread/paper- and paper-based microfluidic devices for glucose assays employing artificial neural networks. Electrophoresis 2018, 39, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Mercan, Ö.B.; Kılıç, V.; Şen, M. Machine learning-based colorimetric determination of glucose in artificial saliva with different reagents using a smartphone coupled μPAD. Sens. Actuators Chem. 2021, 329, 129037. [Google Scholar] [CrossRef]
- Muñoz, H.E.; Riche, C.T.; Kong, J.E.; van Zee, M.; Garner, O.B.; Ozcan, A.; Di Carlo, D. Fractal LAMP: Label-Free Analysis of Fractal Precipitate for Digital Loop-Mediated Isothermal Nucleic Acid Amplification. ACS Sens. 2020, 5, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.; Rahmani, E.; Jew, B.; Garske, K.M.; Miao, Z.; Benhammou, J.N.; Ye, C.J.; Pisegna, J.R.; Pietiläinen, K.H.; Halperin, E.; et al. Enhancing droplet-based single-nucleus RNA-seq resolution using the semi-supervised machine learning classifier DIEM. Sci. Rep. 2020, 10, 11019. [Google Scholar] [CrossRef]
- Lee, T.H.; Kwon, H.B.; Song, W.Y.; Lee, S.S.; Kim, Y.J. Microfluidic ultrafine particle dosimeter using an electrical detection method with a machine-learning-aided algorithm for real-time monitoring of particle density and size distribution. Lab Chip 2021, 21, 1503–1516. [Google Scholar] [CrossRef] [PubMed]
- Alapan, Y.; Kim, C.; Adhikari, A.; Gray, K.E.; Gurkan-Cavusoglu, E.; Little, J.A.; Gurkan, U.A. Sickle cell disease biochip: A functional red blood cell adhesion assay for monitoring sickle cell disease. Transl. Res. 2016, 173, 74–91.e8. [Google Scholar] [CrossRef]
- Praljak, N.; Iram, S.; Goreke, U.; Singh, G.; Hill, A.; Gurkan, U.A.; Hinczewski, M. Integrating deep learning with microfluidics for biophysical classification of sickle red blood cells adhered to laminin. PLoS Comput. Biol. 2021, 17, e1008946. [Google Scholar] [CrossRef]
- Lamoureux, E.S.; Islamzada, E.; Wiens, M.V.J.; Matthews, K.; Duffy, S.P.; Ma, H. Assessing red blood cell deformability from microscopy images using deep learning. Lab Chip 2021, 22, 26–39. [Google Scholar] [CrossRef]
- Rizzuto, V.; Mencattini, A.; Álvarez González, B.; Di Giuseppe, D.; Martinelli, E.; Beneitez-Pastor, D.; Mañú-Pereira, M.d.M.; Lopez-Martinez, M.J.; Samitier, J. Combining microfluidics with machine learning algorithms for RBC classification in rare hereditary hemolytic anemia. Sci. Rep. 2021, 11, 13553. [Google Scholar] [CrossRef]
- Ellett, F.; Jorgensen, J.; Marand, A.L.; Liu, Y.M.; Martinez, M.M.; Sein, V.; Butler, K.L.; Lee, J.; Irimia, D. Diagnosis of sepsis from a drop of blood by measurement of spontaneous neutrophil motility in a microfluidic assay. Nat. Biomed. Eng. 2018, 2, 207–214. [Google Scholar] [CrossRef]
- Kalmady, K.S.; Kamath, A.S.; Gopakumar, G.; Subrahmanyam, G.R.K.S.; Gorthi, S.S. Improved Transfer Learning through Shallow Network Embedding for Classification of Leukemia Cells. In Proceedings of the 2017 Ninth International Conference on Advances in Pattern Recognition (ICAPR), Bangalore, India, 27–30 December 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Gopakumar, G.; Hari Babu, K.; Mishra, D.; Gorthi, S.S.; Sai Subrahmanyam, G.R.K. Cytopathological image analysis using deep-learning networks in microfluidic microscopy. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 2017, 34, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Manak, M.S.; Varsanik, J.S.; Hogan, B.J.; Whitfield, M.J.; Su, W.R.; Joshi, N.; Steinke, N.; Min, A.; Berger, D.; Saphirstein, R.J.; et al. Live-cell phenotypic-biomarker microfluidic assay for the risk stratification of cancer patients via machine learning. Nat. Biomed. Eng. 2018, 2, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Soldati, G.; Del Ben, F.; Brisotto, G.; Biscontin, E.; Bulfoni, M.; Piruska, A.; Steffan, A.; Turetta, M.; Della Mea, V. Microfluidic droplets content classification and analysis through convolutional neural networks in a liquid biopsy workflow. Am. J. Transl. Res. 2018, 10, 4004–4016. [Google Scholar] [PubMed]
- Hashemzadeh, H.; Shojaeilangari, S.; Allahverdi, A.; Rothbauer, M.; Ertl, P.; Naderi-Manesh, H. A combined microfluidic deep learning approach for lung cancer cell high throughput screening toward automatic cancer screening applications. Sci. Rep. 2021, 11, 9804. [Google Scholar] [CrossRef]
- Ayensa-Jiménez, J.; Doweidar, M.H.; Sanz-Herrera, J.A.; Doblare, M. Understanding glioblastoma invasion using physically-guided neural networks with internal variables. PLoS Comput. Biol. 2022, 18, e1010019. [Google Scholar] [CrossRef]
- Ni, W.; Hu, B.; Zheng, C.; Tong, Y.; Wang, L.; Li, Q.q.; Tong, X.; Han, Y. Automated analysis of acute myeloid leukemia minimal residual disease using a support vector machine. Oncotarget 2016, 7, 71915–71921. [Google Scholar] [CrossRef]
- Turan, B.; Masuda, T.; Lei, W.; Noor, A.M.; Horio, K.; Saito, T.I.; Miyata, Y.; Arai, F. A pillar-based microfluidic chip for T-cells and B-cells isolation and detection with machine learning algorithm. ROBOMECH J. 2018, 5, 27. [Google Scholar] [CrossRef]
- McRae, M.P.; Simmons, G.W.; Wong, J.; Shadfan, B.; Gopalkrishnan, S.; Christodoulides, N.; McDevitt, J.T. Programmable bio-nano-chip system: A flexible point-of-care platform for bioscience and clinical measurements. Lab Chip 2015, 15, 4020–4031. [Google Scholar] [CrossRef]
- Christodoulides, N.; De La Garza, R.; Simmons, G.W.; McRae, M.P.; Wong, J.; Newton, T.F.; Smith, R.; Mahoney III, J.J.; Hohenstein, J.; Gomez, S.; et al. Application of programmable bio-nano-chip system for the quantitative detection of drugs of abuse in oral fluids. Drug Alcohol Depend. 2015, 153, 306–313. [Google Scholar] [CrossRef]
- McRae, M.P.; Simmons, G.; Wong, J.; McDevitt, J.T. Programmable Bio-nanochip Platform: A Point-of-Care Biosensor System with the Capacity To Learn. Acc. Chem. Res. 2016, 49, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhao, J.; Cai, T.; Stephens, A.; Su, S.H.; Sandford, E.; Flora, C.; Singer, B.H.; Ghosh, M.; Choi, S.W.; et al. Machine learning-based cytokine microarray digital immunoassay analysis. Biosens. Bioelectron. 2021, 180, 113088. [Google Scholar] [CrossRef] [PubMed]
- Ross, G.M.S.; Filippini, D.; Nielen, M.W.F.; Salentijn, G.I. Unraveling the Hook Effect: A Comprehensive Study of High Antigen Concentration Effects in Sandwich Lateral Flow Immunoassays. Anal. Chem. 2020, 92, 15587–15595. [Google Scholar] [CrossRef] [PubMed]
- Banaei, N.; Moshfegh, J.; Mohseni-Kabir, A.; Houghton, J.M.; Sun, Y.; Kim, B. Machine learning algorithms enhance the specificity of cancer biomarker detection using SERS-based immunoassays in microfluidic chips. RSC Adv. 2019, 9, 1859–1868. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Joung, H.A.; Esparza, S.; Rao, J.; Garner, O.; Ozcan, A. Quantitative particle agglutination assay for point-of-care testing using mobile holographic imaging and deep learning. Lab Chip 2021, 21, 3550–3558. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Kim, S.; Hardie, J.M.; Thirumalaraju, P.; Gharpure, S.; Rostamian, S.; Udayakumar, S.; Lei, Q.; Cho, G.; Kanakasabapathy, M.K.; et al. Deep learning-assisted sensitive detection of fentanyl using a bubbling-microchip. Lab Chip 2022, 22, 4531–4540. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Khan, S.; Tiwari, R.; Sircar, S.; Bhat, S.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Coronavirus Disease 2019–COVID-19. Clin. Microbiol. Rev. 2020, 33, e00028-20. [Google Scholar] [CrossRef]
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, 782–793. [Google Scholar] [CrossRef]
- Funari, R.; Chu, K.Y.; Shen, A.Q. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens. Bioelectron. 2020, 169, 112578. [Google Scholar] [CrossRef]
- Jamiruddin, M.R.; Meghla, B.A.; Islam, D.Z.; Tisha, T.A.; Khandker, S.S.; Khondoker, M.U.; Haq, M.A.; Adnan, N.; Haque, M. Microfluidics Technology in SARS-CoV-2 Diagnosis and Beyond: A Systematic Review. Life 2022, 12, 649. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, X.; Wang, Q.; Liu, W.; Chen, C. Microfluidics for COVID-19: From Current Work to Future Perspective. Biosensors 2023, 13, 163. [Google Scholar] [CrossRef]
- Bhuiyan, N.H.; Hong, J.H.; Uddin, M.J.; Shim, J.S. Artificial Intelligence-Controlled Microfluidic Device for Fluid Automation and Bubble Removal of Immunoassay Operated by a Smartphone. Anal. Chem. 2022, 94, 3872–3880. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, N.H.; Uddin, M.J.; Lee, J.; Hong, J.H.; Shim, J.S. An Internet-of-Disease System for COVID-19 Testing Using Saliva by an AI-Controlled Microfluidic ELISA Device. Adv. Mater. Technol. 2022, 7, 2101690. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Fu, Y.Q.; Jin, N.; Chazot, P.; Angelov, P.; Jiang, R. AI-enabled Microscopic Blood Analysis for Microfluidic COVID-19 Hematology. In Proceedings of the 2020 5th International Conference on Computational Intelligence and Applications (ICCIA), Beijing, China, 19–21 June 2020; pp. 98–102. [Google Scholar] [CrossRef]
- Gao, Z.; Song, Y.; Hsiao, T.Y.; He, J.; Wang, C.; Shen, J.; MacLachlan, A.; Dai, S.; Singer, B.H.; Kurabayashi, K.; et al. Machine-Learning-Assisted Microfluidic Nanoplasmonic Digital Immunoassay for Cytokine Storm Profiling in COVID-19 Patients. ACS Nano 2021, 15, 18023–18036. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.J.; Hu, Y.; Xiong, Z.; Wang, J.; McLeod, E. Point-of-care SARS-CoV-2 sensing using lens-free imaging and a deep learning-assisted quantitative agglutination assay. Lab Chip 2022, 22, 3744–3754. [Google Scholar] [CrossRef]
- Wang, B.; Li, Y.; Zhou, M.; Han, Y.; Zhang, M.; Gao, Z.; Liu, Z.; Chen, P.; Du, W.; Zhang, X.; et al. Smartphone-based platforms implementing microfluidic detection with image-based artificial intelligence. Nat. Commun. 2023, 14, 1341. [Google Scholar] [CrossRef]
- Kumar, A.; Parihar, A.; Panda, U.; Parihar, D.S. Microfluidics-Based Point-of-Care Testing (POCT) Devices in Dealing with Waves of COVID-19 Pandemic: The Emerging Solution. ACS Appl. Bio Mater. 2022, 5, 2046–2068. [Google Scholar] [CrossRef]
- Ramezankhani, R.; Solhi, R.; Chai, Y.C.; Vosough, M.; Verfaillie, C. Organoid and microfluidics-based platforms for drug screening in COVID-19. Drug Discov. Today 2022, 27, 1062–1076. [Google Scholar] [CrossRef]
- Chiu, H.Y.R.; Hwang, C.K.; Chen, S.Y.; Shih, F.Y.; Han, H.C.; King, C.C.; Gilbert, J.R.; Fang, C.C.; Oyang, Y.J. Machine learning for emerging infectious disease field responses. Sci. Rep. 2022, 12, 328. [Google Scholar] [CrossRef]
- Tran, N.K.; Albahra, S.; May, L.; Waldman, S.; Crabtree, S.; Bainbridge, S.; Rashidi, H. Evolving Applications of Artificial Intelligence and Machine Learning in Infectious Diseases Testing. Clin. Chem. 2022, 68, 125–133. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, L.; Wang, Y.; Zhang, T.; Chen, Y.C.; Yoon, E. Label-Free Estimation of Therapeutic Efficacy on 3D Cancer Spheres Using Convolutional Neural Network Image Analysis. Anal. Chem. 2019, 91, 14093–14100. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Hu, S.; Song, W.; Gu, M.; Liu, J.; Song, J.; Liu, Z.; Li, Z.; Huang, K.; Wu, Y.; et al. An ultralight, flexible, and biocompatible all-fiber motion sensor for artificial intelligence wearable electronics. Npj Flex. Electron. 2022, 6, 1–8. [Google Scholar] [CrossRef]
- Kobayashi, H.; Lei, C.; Wu, Y.; Mao, A.; Jiang, Y.; Guo, B.; Ozeki, Y.; Goda, K. Label-free detection of cellular drug responses by high-throughput bright-field imaging and machine learning. Sci. Rep. 2017, 7, 12454. [Google Scholar] [CrossRef]
- Kobayashi, H.; Lei, C.; Wu, Y.; Huang, C.J.; Yasumoto, A.; Jona, M.; Li, W.; Wu, Y.; Yalikun, Y.; Jiang, Y.; et al. Intelligent whole-blood imaging flow cytometry for simple, rapid, and cost-effective drug-susceptibility testing of leukemia. Lab Chip 2019, 19, 2688–2698. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yasumoto, A.; Lei, C.; Huang, C.J.; Kobayashi, H.; Wu, Y.; Yan, S.; Sun, C.W.; Yatomi, Y.; Goda, K. Intelligent classification of platelet aggregates by agonist type. eLife 2020, 9, e52938. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Deng, S.; Zhang, L. A review of artificial intelligence applications for antimicrobial resistance. Biosaf. Health 2021, 3, 22–31. [Google Scholar] [CrossRef]
- Poweleit, E.A.; Vinks, A.A.; Mizuno, T. Artificial Intelligence and Machine Learning Approaches to Facilitate Therapeutic Drug Management and Model-Informed Precision Dosing. Ther. Drug Monit. 2022, 45, 143–150. [Google Scholar] [CrossRef]
- Yu, H.; Jing, W.; Iriya, R.; Yang, Y.; Syal, K.; Mo, M.; Grys, T.E.; Haydel, S.E.; Wang, S.; Tao, N. Phenotypic Antimicrobial Susceptibility Testing with Deep Learning Video Microscopy. Anal. Chem. 2018, 90, 6314–6322. [Google Scholar] [CrossRef]
- Kim, K.; Kim, S.; Jeon, J.S. Visual Estimation of Bacterial Growth Level in Microfluidic Culture Systems. Sensors 2018, 18, 447. [Google Scholar] [CrossRef]
- Svensson, C.M.; Shvydkiv, O.; Dietrich, S.; Mahler, L.; Weber, T.; Choudhary, M.; Tovar, M.; Figge, M.T.; Roth, M. Coding of Experimental Conditions in Microfluidic Droplet Assays Using Colored Beads and Machine Learning Supported Image Analysis. Small 2019, 15, 1802384. [Google Scholar] [CrossRef]
- Rauf, S.; Tashkandi, N.; de Oliveira Filho, J.I.; Oviedo-Osornio, C.I.; Danish, M.S.; Hong, P.Y.; Salama, K.N. Digital E. coli Counter: A Microfluidics and Computer Vision-Based DNAzyme Method for the Isolation and Specific Detection of E. coli from Water Samples. Biosensors 2022, 12, 34. [Google Scholar] [CrossRef] [PubMed]
- Alves, V.M.; Auerbach, S.S.; Kleinstreuer, N.; Rooney, J.P.; Muratov, E.N.; Rusyn, I.; Tropsha, A.; Schmitt, C. Curated Data In—Trustworthy In Silico Models Out: The Impact of Data Quality on the Reliability of Artificial Intelligence Models as Alternatives to Animal Testing. Altern. Lab. Anim. 2021, 49, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tao, T.H. Skin-Friendly Electronics for Acquiring Human Physiological Signatures. Adv. Mater. 2019, 31, 1905767. [Google Scholar] [CrossRef]
- Huang, J.D.; Wang, J.; Ramsey, E.; Leavey, G.; Chico, T.J.A.; Condell, J. Applying Artificial Intelligence to Wearable Sensor Data to Diagnose and Predict Cardiovascular Disease: A Review. Sensors 2022, 22, 8002. [Google Scholar] [CrossRef]
- Rodríguez-Rodríguez, I.; Rodríguez, J.V.; Chatzigiannakis, I.; Zamora Izquierdo, M.A. On the Possibility of Predicting Glycaemia ‘On the Fly’ with Constrained IoT Devices in Type 1 Diabetes Mellitus Patients. Sensors 2019, 19, 4538. [Google Scholar] [CrossRef] [PubMed]
- Sankhala, D.; Sardesai, A.U.; Pali, M.; Lin, K.C.; Jagannath, B.; Muthukumar, S.; Prasad, S. A machine learning-based on-demand sweat glucose reporting platform. Sci. Rep. 2022, 12, 2442. [Google Scholar] [CrossRef]
- Han, S.; Kim, T.; Kim, D.; Park, Y.L.; Jo, S. Use of Deep Learning for Characterization of Microfluidic Soft Sensors. IEEE Robot. Autom. Lett. 2018, 3, 873–880. [Google Scholar] [CrossRef]
- Kim, D.; Kim, M.; Kwon, J.; Park, Y.L.; Jo, S. Semi-Supervised Gait Generation with Two Microfluidic Soft Sensors. IEEE Robot. Autom. Lett. 2019, 4, 2501–2507. [Google Scholar] [CrossRef]
- Wang, Y.; Shan, G.; Li, H.; Wang, L. A Wearable-Sensor System with AI Technology for Real-Time Biomechanical Feedback Training in Hammer Throw. Sensors 2023, 23, 425. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, Y.; Jiang, N.; Yetisen, A.K. Wearable artificial intelligence biosensor networks. Biosens. Bioelectron. 2023, 219, 114825. [Google Scholar] [CrossRef]
- Helmy, M.; Smith, D.; Selvarajoo, K. Systems biology approaches integrated with artificial intelligence for optimized metabolic engineering. Metab. Eng. Commun. 2020, 11, e00149. [Google Scholar] [CrossRef] [PubMed]
- Riordon, J.; Sovilj, D.; Sanner, S.; Sinton, D.; Young, E.W.K. Deep Learning with Microfluidics for Biotechnology. Trends Biotechnol. 2019, 37, 310–324. [Google Scholar] [CrossRef]
- Raji, H.; Tayyab, M.; Sui, J.; Mahmoodi, S.R.; Javanmard, M. Biosensors and machine learning for enhanced detection, stratification, and classification of cells: A review. Biomed. Microdevices 2022, 24, 26. [Google Scholar] [CrossRef]
- Yang Yu, B.; Elbuken, C.; Ren, C.L.; Huissoon, J.P. Image processing and classification algorithm for yeast cell morphology in a microfluidic chip. J. Biomed. Opt. 2011, 16, 066008. [Google Scholar] [CrossRef]
- Huang, X.; Guo, J.; Wang, X.; Yan, M.; Kang, Y.; Yu, H. A Contact-Imaging Based Microfluidic Cytometer with Machine-Learning for Single-Frame Super-Resolution Processing. PLoS ONE 2014, 9, e104539. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, R.; Asmare, N.; Chu, C.H.; Sarioglu, A.F. Processing code-multiplexed Coulter signals via deep convolutional neural networks. Lab Chip 2019, 19, 3292–3304. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zheng, Y.; Tan, Q.; Shojaei-Baghini, E.; Zhang, Y.L.; Li, J.; Prasad, P.; You, L.; Wu, X.Y.; Sun, Y. Classification of cell types using a microfluidic device for mechanical and electrical measurement on single cells. Lab Chip 2011, 11, 3174–3181. [Google Scholar] [CrossRef]
- Heo, Y.J.; Lee, D.; Kang, J.; Lee, K.; Chung, W.K. Real-time Image Processing for Microscopy-based Label-free Imaging Flow Cytometry in a Microfluidic Chip. Sci. Rep. 2017, 7, 11651. [Google Scholar] [CrossRef]
- Constantinou, I.; Jendrusch, M.; Aspert, T.; Görlitz, F.; Schulze, A.; Charvin, G.; Knop, M. Self-Learning Microfluidic Platform for Single-Cell Imaging and Classification in Flow. Micromachines 2019, 10, 311. [Google Scholar] [CrossRef]
- Mikami, H.; Kawaguchi, M.; Huang, C.J.; Matsumura, H.; Sugimura, T.; Huang, K.; Lei, C.; Ueno, S.; Miura, T.; Ito, T.; et al. Virtual-freezing fluorescence imaging flow cytometry. Nat. Commun. 2020, 11, 1162. [Google Scholar] [CrossRef]
- Huang, K.; Matsumura, H.; Zhao, Y.; Herbig, M.; Yuan, D.; Mineharu, Y.; Harmon, J.; Findinier, J.; Yamagishi, M.; Ohnuki, S.; et al. Deep imaging flow cytometry. Lab Chip 2022, 22, 876–889. [Google Scholar] [CrossRef]
- Ahmad, A.; Sala, F.; Paiè, P.; Candeo, A.; D’Annunzio, S.; Zippo, A.; Frindel, C.; Osellame, R.; Bragheri, F.; Bassi, A.; et al. On the robustness of machine learning algorithms toward microfluidic distortions for cell classification via on-chip fluorescence microscopy. Lab Chip 2022, 22, 3453–3463. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Dannhauser, D.; Telesco, M.; Netti, P.A.; Causa, F. CD4+ versus CD8+ T-lymphocyte identification in an integrated microfluidic chip using light scattering and machine learning. Lab Chip 2019, 19, 3888–3898. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.; Meng, J.; Ding, G.; Shi, Z.; Wang, R.; Zhang, X. Detection of non-small cell lung cancer cells based on microfluidic polarization microscopic image analysis. Electrophoresis 2019, 40, 1202–1211. [Google Scholar] [CrossRef]
- Singh, D.K.; Ahrens, C.C.; Li, W.; Vanapalli, S.A. Label-free, high-throughput holographic screening and enumeration of tumor cells in blood. Lab Chip 2017, 17, 2920–2932. [Google Scholar] [CrossRef] [PubMed]
- Feizi, A.; Zhang, Y.; Greenbaum, A.; Guziak, A.; Luong, M.; Chan, R.Y.L.; Berg, B.; Ozkan, H.; Luo, W.; Wu, M.; et al. Rapid, portable and cost-effective yeast cell viability and concentration analysis using lensfree on-chip microscopy and machine learning. Lab Chip 2016, 16, 4350–4358. [Google Scholar] [CrossRef]
- Nissim, N.; Dudaie, M.; Barnea, I.; Shaked, N.T. Real-Time Stain-Free Classification of Cancer Cells and Blood Cells Using Interferometric Phase Microscopy and Machine Learning. Cytom. Part A 2021, 99, 511–523. [Google Scholar] [CrossRef]
- Hirotsu, A.; Kikuchi, H.; Yamada, H.; Ozaki, Y.; Haneda, R.; Kawata, S.; Murakami, T.; Matsumoto, T.; Hiramatsu, Y.; Kamiya, K.; et al. Artificial intelligence-based classification of peripheral blood nucleated cells using label-free imaging flow cytometry. Lab Chip 2022, 22, 3464–3474. [Google Scholar] [CrossRef]
- Wu, J.L.; Xu, Y.Q.; Xu, J.J.; Wei, X.M.; Chan, A.C.; Tang, A.H.; Lau, A.K.; Chung, B.M.; Cheung Shum, H.; Lam, E.Y.; et al. Ultrafast laser-scanning time-stretch imaging at visible wavelengths. Light. Sci. Appl. 2017, 6, e16196. [Google Scholar] [CrossRef]
- Pirone, D.; Sirico, D.; Miccio, L.; Bianco, V.; Mugnano, M.; Ferraro, P.; Memmolo, P. Speeding up reconstruction of 3D tomograms in holographic flow cytometry via deep learning. Lab Chip 2022, 22, 793–804. [Google Scholar] [CrossRef]
- Jiang, Y.; Lei, C.; Yasumoto, A.; Kobayashi, H.; Aisaka, Y.; Ito, T.; Guo, B.; Nitta, N.; Kutsuna, N.; Ozeki, Y.; et al. Label-free detection of aggregated platelets in blood by machine-learning-aided optofluidic time-stretch microscopy. Lab Chip 2017, 17, 2426–2434. [Google Scholar] [CrossRef]
- Guo, B.; Lei, C.; Kobayashi, H.; Ito, T.; Yalikun, Y.; Jiang, Y.; Tanaka, Y.; Ozeki, Y.; Goda, K. High-throughput, label-free, single-cell, microalgal lipid screening by machine-learning-equipped optofluidic time-stretch quantitative phase microscopy. Cytom. Part A 2017, 91, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Mahjoubfar, A.; Chen, C.L.; Niazi, K.R.; Pei, L.; Jalali, B. Deep Cytometry: Deep learning with Real-time Inference in Cell Sorting and Flow Cytometry. Sci. Rep. 2019, 9, 11088. [Google Scholar] [CrossRef]
- Lee, K.C.; Wang, M.; Cheah, K.S.; Chan, G.C.; So, H.K.; Wong, K.K.; Tsia, K.K. Quantitative Phase Imaging Flow Cytometry for Ultra-Large-Scale Single-Cell Biophysical Phenotyping. Cytom. Part A 2019, 95, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Yip, G.G.K.; Lo, M.C.K.; Yan, W.; Lee, K.C.M.; Lai, Q.T.K.; Wong, K.K.Y.; Tsia, K.K. Multimodal FACED imaging for large-scale single-cell morphological profiling. APL Photonics 2021, 6, 070801. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, Y.; Huang, C.J.; Huang, C.J.; Kobayashi, H.; Kobayashi, H.; Yan, S.; Ozeki, Y.; Wu, Y.; Sun, C.W.; et al. Intelligent frequency-shifted optofluidic time-stretch quantitative phase imaging. Opt. Express 2020, 28, 519–532. [Google Scholar] [CrossRef]
- Joshi, K.; Javani, A.; Park, J.; Velasco, V.; Xu, B.; Razorenova, O.; Esfandyarpour, R. A Machine Learning-Assisted Nanoparticle-Printed Biochip for Real-Time Single Cancer Cell Analysis. Adv. Biosyst. 2020, 4, 2000160. [Google Scholar] [CrossRef]
- Honrado, C.; McGrath, J.S.; Reale, R.; Bisegna, P.; Swami, N.S.; Caselli, F. A neural network approach for real-time particle/cell characterization in microfluidic impedance cytometry. Anal. Bioanal. Chem. 2020, 412, 3835–3845. [Google Scholar] [CrossRef]
- Wang, N.; Liu, R.; Asmare, N.; Chu, C.H.; Civelekoglu, O.; Sarioglu, A.F. Closed-loop feedback control of microfluidic cell manipulation via deep-learning integrated sensor networks. Lab Chip 2021, 21, 1916–1928. [Google Scholar] [CrossRef]
- Feng, Y.; Cheng, Z.; Chai, H.; He, W.; Huang, L.; Wang, W. Neural network-enhanced real-time impedance flow cytometry for single-cell intrinsic characterization. Lab Chip 2022, 22, 240–249. [Google Scholar] [CrossRef]
- Caselli, F.; Reale, R.; Ninno, A.D.; Spencer, D.; Morgan, H.; Bisegna, P. Deciphering impedance cytometry signals with neural networks. Lab Chip 2022, 22, 1714–1722. [Google Scholar] [CrossRef]
- Robinson, J.P. Flow cytometry: Past and future. BioTechniques 2022, 72, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhang, A.C.; Han, Y.; Li, J.; Chen, C.; Lo, Y.H. Machine Learning Based Real-Time Image-Guided Cell Sorting and Classification. Cytometry. Part A J. Int. Soc. Anal. Cytol. 2019, 95, 499–509. [Google Scholar] [CrossRef]
- Sesen, M.; Whyte, G. Image-Based Single Cell Sorting Automation in Droplet Microfluidics. Sci. Rep. 2020, 10, 8736. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kim, S.E.; Doh, J.; Kim, K.; Chung, W.K. User-friendly image-activated microfluidic cell sorting technique using an optimized, fast deep learning algorithm. Lab Chip 2021, 21, 1798–1810. [Google Scholar] [CrossRef]
- Nitta, N.; Sugimura, T.; Isozaki, A.; Mikami, H.; Hiraki, K.; Sakuma, S.; Iino, T.; Arai, F.; Endo, T.; Fujiwaki, Y.; et al. Intelligent Image-Activated Cell Sorting. Cell 2018, 175, 266–276.e13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ouyang, M.; Ray, A.; Liu, T.; Kong, J.; Bai, B.; Kim, D.; Guziak, A.; Luo, Y.; Feizi, A.; et al. Computational cytometer based on magnetically modulated coherent imaging and deep learning. Light. Sci. Appl. 2019, 8, 91. [Google Scholar] [CrossRef] [PubMed]
- Isozaki, A.; Mikami, H.; Tezuka, H.; Matsumura, H.; Huang, K.; Akamine, M.; Hiramatsu, K.; Iino, T.; Ito, T.; Karakawa, H.; et al. Intelligent image-activated cell sorting 2.0. Lab Chip 2020, 20, 2263–2273. [Google Scholar] [CrossRef]
- McCallum, C.; Riordon, J.; Wang, Y.; Kong, T.; You, J.B.; Sanner, S.; Lagunov, A.; Hannam, T.G.; Jarvi, K.; Sinton, D. Deep learning-based selection of human sperm with high DNA integrity. Commun. Biol. 2019, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Uslu, F.; Icoz, K.; Tasdemir, K.; Yilmaz, B. Automated quantification of immunomagnetic beads and leukemia cells from optical microscope images. Biomed. Signal Process. Control 2019, 49, 473–482. [Google Scholar] [CrossRef]
- White, A.M.; Zhang, Y.; Shamul, J.G.; Xu, J.; Kwizera, E.A.; Jiang, B.; He, X. Deep Learning-Enabled Label-Free On-Chip Detection and Selective Extraction of Cell Aggregate-Laden Hydrogel Microcapsules. Small 2021, 17, 2100491. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, E.M.; Yang, S.J.; Ando, D.M.; Javaherian, A.; Skibinski, G.; Lipnick, S.; Mount, E.; O’Neil, A.; Shah, K.; Lee, A.K.; et al. In Silico Labeling: Predicting Fluorescent Labels in Unlabeled Images. Cell 2018, 173, 792–803.e19. [Google Scholar] [CrossRef]
- Ounkomol, C.; Seshamani, S.; Maleckar, M.M.; Collman, F.; Johnson, G.R. Label-free prediction of three-dimensional fluorescence images from transmitted-light microscopy. Nat. Methods 2018, 15, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Min, Y.; Oh, J.M.; Cho, Y.K. AI-powered transmitted light microscopy for functional analysis of live cells. Sci. Rep. 2019, 9, 18428. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Yang, X.; Li, Y.; Zhang, Y.; Pillar, N.; Ozcan, A. Deep learning-enabled virtual histological staining of biological samples. Light. Sci. Appl. 2023, 12, 57. [Google Scholar] [CrossRef]
- Ugawa, M.; Kawamura, Y.; Toda, K.; Teranishi, K.; Morita, H.; Adachi, H.; Tamoto, R.; Nomaru, H.; Nakagawa, K.; Sugimoto, K.; et al. In silico-labeled ghost cytometry. eLife 2021, 10, e67660. [Google Scholar] [CrossRef]
- Yang, D.; Zhou, Y.; Zhou, Y.; Han, J.; Ai, Y. Biophysical phenotyping of single cells using a differential multiconstriction microfluidic device with self-aligned 3D electrodes. Biosens. Bioelectron. 2019, 133, 16–23. [Google Scholar] [CrossRef]
- Xiao, W.; Xin, L.; Cao, R.; Wu, X.; Tian, R.; Che, L.; Sun, L.; Ferraro, P.; Pan, F. Sensing morphogenesis of bone cells under microfluidic shear stress by holographic microscopy and automatic aberration compensation with deep learning. Lab Chip 2021, 21, 1385–1394. [Google Scholar] [CrossRef]
- Soelistyo, C.J.; Vallardi, G.; Charras, G.; Lowe, A.R. Learning biophysical determinants of cell fate with deep neural networks. Nat. Mach. Intell. 2022, 4, 636–644. [Google Scholar] [CrossRef]
- Combs, C.; Seith, D.D.; Bovyn, M.J.; Gross, S.P.; Xie, X.; Siwy, Z.S. Deep learning assisted mechanotyping of individual cells through repeated deformations and relaxations in undulating channels. Biomicrofluidics 2022, 16, 014104. [Google Scholar] [CrossRef]
- Sarkar, S.; Kang, W.; Jiang, S.; Li, K.; Ray, S.; Luther, E.; Ivanov, A.R.; Fu, Y.; Konry, T. Machine learning-aided quantification of antibody-based cancer immunotherapy by natural killer cells in microfluidic droplets. Lab Chip 2020, 20, 2317–2327. [Google Scholar] [CrossRef] [PubMed]
- Ao, Z.; Cai, H.; Wu, Z.; Hu, L.; Nunez, A.; Zhou, Z.; Liu, H.; Bondesson, M.; Lu, X.; Lu, X.; et al. Microfluidics guided by deep learning for cancer immunotherapy screening. Proc. Natl. Acad. Sci. 2022, 119, e2214569119. [Google Scholar] [CrossRef] [PubMed]
- Honrado, C.; Salahi, A.; Adair, S.J.; Moore, J.H.; Bauer, T.W.; Swami, N.S. Automated biophysical classification of apoptotic pancreatic cancer cell subpopulations by using machine learning approaches with impedance cytometry. Lab Chip 2022, 22, 3708–3720. [Google Scholar] [CrossRef]
- Whitfield, M.L.; Sherlock, G.; Saldanha, A.J.; Murray, J.I.; Ball, C.A.; Alexander, K.E.; Matese, J.C.; Perou, C.M.; Hurt, M.M.; Brown, P.O.; et al. Identification of Genes Periodically Expressed in the Human Cell Cycle and Their Expression in Tumors. Mol. Biol. Cell 2002, 13, 1977–2000. [Google Scholar] [CrossRef]
- Song, H.; Wang, Y.; Rosano, J.M.; Prabhakarpandian, B.; Garson, C.; Pant, K.; Lai, E. A microfluidic impedance flow cytometer for identification of differentiation state of stem cells. Lab Chip 2013, 13, 2300–2310. [Google Scholar] [CrossRef] [PubMed]
- Eulenberg, P.; Köhler, N.; Blasi, T.; Filby, A.; Carpenter, A.E.; Rees, P.; Theis, F.J.; Wolf, F.A. Reconstructing cell cycle and disease progression using deep learning. Nat. Commun. 2017, 8, 463. [Google Scholar] [CrossRef]
- Rappez, L.; Rakhlin, A.; Rigopoulos, A.; Nikolenko, S.; Alexandrov, T. DeepCycle reconstructs a cyclic cell cycle trajectory from unsegmented cell images using convolutional neural networks. Mol. Syst. Biol. 2020, 16, e9474. [Google Scholar] [CrossRef]
- He, Y.R.; He, S.; Kandel, M.E.; Lee, Y.J.; Hu, C.; Sobh, N.; Anastasio, M.A.; Popescu, G. Cell Cycle Stage Classification Using Phase Imaging with Computational Specificity. ACS Photonics 2022, 9, 1264–1273. [Google Scholar] [CrossRef]
- Ghafari, M.; Clark, J.; Guo, H.B.; Yu, R.; Sun, Y.; Dang, W.; Qin, H. Complementary performances of convolutional and capsule neural networks on classifying microfluidic images of dividing yeast cells. PLoS ONE 2021, 16, e0246988. [Google Scholar] [CrossRef]
- Aspert, T.; Hentsch, D.; Charvin, G. DetecDiv, a generalist deep-learning platform for automated cell division tracking and survival analysis. eLife 2022, 11, e79519. [Google Scholar] [CrossRef]
- Kusumoto, D.; Seki, T.; Sawada, H.; Kunitomi, A.; Katsuki, T.; Kimura, M.; Ito, S.; Komuro, J.; Hashimoto, H.; Fukuda, K.; et al. Anti-senescent drug screening by deep learning-based morphology senescence scoring. Nat. Commun. 2021, 12, 257. [Google Scholar] [CrossRef] [PubMed]
- Heckenbach, I.; Mkrtchyan, G.V.; Ezra, M.B.; Bakula, D.; Madsen, J.S.; Nielsen, M.H.; Oró, D.; Osborne, B.; Covarrubias, A.J.; Idda, M.L.; et al. Nuclear morphology is a deep learning biomarker of cellular senescence. Nat. Aging 2022, 2, 742–755. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015. Lecture Notes in Computer Science. pp. 234–241. [Google Scholar] [CrossRef]
- Valen, D.A.V.; Kudo, T.; Lane, K.M.; Macklin, D.N.; Quach, N.T.; DeFelice, M.M.; Maayan, I.; Tanouchi, Y.; Ashley, E.A.; Covert, M.W. Deep Learning Automates the Quantitative Analysis of Individual Cells in Live-Cell Imaging Experiments. PLoS Comput. Biol. 2016, 12, e1005177. [Google Scholar] [CrossRef]
- Tsai, H.F.; Gajda, J.; Sloan, T.F.W.; Rares, A.; Shen, A.Q. Usiigaci: Instance-aware cell tracking in stain-free phase contrast microscopy enabled by machine learning. SoftwareX 2019, 9, 230–237. [Google Scholar] [CrossRef]
- Bove, A.; Gradeci, D.; Fujita, Y.; Banerjee, S.; Charras, G.; Lowe, A.R. Local cellular neighborhood controls proliferation in cell competition. Mol. Biol. Cell 2017, 28, 3215–3228. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Miura, T.; Voleti, V.; Yamaguchi, K.; Tsutsumi, M.; Yamamoto, K.; Otomo, K.; Fujie, Y.; Teramoto, T.; Ishihara, T.; et al. 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images. eLife 2021, 10, e59187. [Google Scholar] [CrossRef]
- Wang, A.; Zhang, Q.; Han, Y.; Megason, S.; Hormoz, S.; Mosaliganti, K.R.; Lam, J.C.K.; Li, V.O.K. A novel deep learning-based 3D cell segmentation framework for future image-based disease detection. Sci. Rep. 2022, 12, 342. [Google Scholar] [CrossRef]
- Padovani, F.; Mairhörmann, B.; Falter-Braun, P.; Lengefeld, J.; Schmoller, K.M. Segmentation, tracking and cell cycle analysis of live-cell imaging data with Cell-ACDC. BMC Biol. 2022, 20, 174. [Google Scholar] [CrossRef]
- Ershov, D.; Phan, M.S.; Pylvänäinen, J.W.; Rigaud, S.U.; Le Blanc, L.; Charles-Orszag, A.; Conway, J.R.W.; Laine, R.F.; Roy, N.H.; Bonazzi, D.; et al. TrackMate 7: Integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods 2022, 19, 829–832. [Google Scholar] [CrossRef]
- Alnahhas, R.N.; Winkle, J.J.; Hirning, A.J.; Karamched, B.; Ott, W.; Josić, K.; Bennett, M.R. Spatiotemporal Dynamics of Synthetic Microbial Consortia in Microfluidic Devices. ACS Synth. Biol. 2019, 8, 2051–2058. [Google Scholar] [CrossRef]
- Lugagne, J.B.; Lin, H.; Dunlop, M.J. DeLTA: Automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Comput. Biol. 2020, 16, e1007673. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, O.M.; Alnahhas, R.N.; Lugagne, J.B.; Dunlop, M.J. DeLTA 2.0: A deep learning pipeline for quantifying single-cell spatial and temporal dynamics. PLoS Comput. Biol. 2022, 18, e1009797. [Google Scholar] [CrossRef] [PubMed]
- Koldaeva, A.; Tsai, H.F.; Shen, A.Q.; Pigolotti, S. Population genetics in microchannels. Proc. Natl. Acad. Sci. 2022, 119, e2120821119. [Google Scholar] [CrossRef]
- Ulicna, K.; Vallardi, G.; Charras, G.; Lowe, A.R. Automated Deep Lineage Tree Analysis Using a Bayesian Single Cell Tracking Approach. Front. Comput. Sci. 2021, 3, 734559. [Google Scholar] [CrossRef]
- Wang, M.; Ong, L.L.S.; Dauwels, J.; Asada, H.H. Multicell migration tracking within angiogenic networks by deep learning-based segmentation and augmented Bayesian filtering. J. Med. Imaging 2018, 5, 024005. [Google Scholar] [CrossRef]
- Tsai, H.F.; IJspeert, C.; Shen, A.Q. Voltage-gated ion channels mediate the electrotaxis of glioblastoma cells in a hybrid PMMA/PDMS microdevice. APL Bioeng. 2020, 4, 036102. [Google Scholar] [CrossRef]
- Stallmann, D.; Göpfert, J.P.; Schmitz, J.; Grünberger, A.; Hammer, B. Towards an automatic analysis of CHO-K1 suspension growth in microfluidic single-cell cultivation. Bioinformatics 2021, 37, 3632–3639. [Google Scholar] [CrossRef] [PubMed]
- Kok, R.N.U.; Hebert, L.; Huelsz-Prince, G.; Goos, Y.J.; Zheng, X.; Bozek, K.; Stephens, G.J.; Tans, S.J.; Zon, J.S.v. OrganoidTracker: Efficient cell tracking using machine learning and manual error correction. PLoS ONE 2020, 15, e0240802. [Google Scholar] [CrossRef]
- Sugawara, K.; Çevrim, Ç.; Averof, M. Tracking cell lineages in 3D by incremental deep learning. eLife 2022, 11, e69380. [Google Scholar] [CrossRef]
- Johnson, K.B.; Wei, W.Q.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision Medicine, AI, and the Future of Personalized Health Care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef]
- Guo, S.C.; Tao, S.C.; Dawn, H. Microfluidics-based on-a-chip systems for isolating and analysing extracellular vesicles. J. Extracell. Vesicles 2018, 7, 1508271. [Google Scholar] [CrossRef]
- Ayuso, J.M.; Virumbrales-Muñoz, M.; Lang, J.M.; Beebe, D.J. A role for microfluidic systems in precision medicine. Nat. Commun. 2022, 13, 3086. [Google Scholar] [CrossRef]
- Van Norman, G.A. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Part 2: Potential Alternatives to the Use of Animals in Preclinical Trials. JACC Basic Transl. Sci. 2020, 5, 387–397. [Google Scholar] [CrossRef] [PubMed]
- van Berlo, D.; van de Steeg, E.; Amirabadi, H.E.; Masereeuw, R. The potential of multi-organ-on-chip models for assessment of drug disposition as alternative to animal testing. Curr. Opin. Toxicol. 2021, 27, 8–17. [Google Scholar] [CrossRef]
- Ma, S.; Murphy, T.W.; Lu, C. Microfluidics for genome-wide studies involving next generation sequencing. Biomicrofluidics 2017, 11, 021501. [Google Scholar] [CrossRef]
- Zhou, W.m.; Yan, Y.y.; Guo, Q.r.; Ji, H.; Wang, H.; Xu, T.t.; Makabel, B.; Pilarsky, C.; He, G.; Yu, X.y.; et al. Microfluidics applications for high-throughput single cell sequencing. J. Nanobiotechnol. 2021, 19, 312. [Google Scholar] [CrossRef] [PubMed]
- Lamanna, J.; Scott, E.Y.; Edwards, H.S.; Chamberlain, M.D.; Dryden, M.D.M.; Peng, J.; Mair, B.; Lee, A.; Chan, C.; Sklavounos, A.A.; et al. Digital microfluidic isolation of single cells for -Omics. Nat. Commun. 2020, 11, 5632. [Google Scholar] [CrossRef]
- Heydari, A.A.; Sindi, S.S. Deep learning in spatial transcriptomics: Learning from the next next-generation sequencing. Biophys. Rev. 2023, 4, 011306. [Google Scholar] [CrossRef]
- Ko, J.; Bhagwat, N.; Yee, S.S.; Ortiz, N.; Sahmoud, A.; Black, T.; Aiello, N.M.; McKenzie, L.; O’Hara, M.; Redlinger, C.; et al. Combining Machine Learning and Nanofluidic Technology To Diagnose Pancreatic Cancer Using Exosomes. ACS Nano 2017, 11, 11182–11193. [Google Scholar] [CrossRef]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2021, 20, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Guo, Y.; Zhu, Y.; Qin, J. Engineering stem cell-derived 3D brain organoids in a perfusable organ-on-a-chip system. RSC Adv. 2018, 8, 1677–1685. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.G.; Jeong, Y.H.; Kim, Y.; Choi, Y.J.; Moon, H.E.; Park, S.H.; Kang, K.S.; Bae, M.; Jang, J.; Youn, H.; et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, D.J.; Kip, A.M.; Romitti, M.; Nazzari, M.; Tegel, A.; Stich, M.; Krause, C.; Caiment, F.; Costagliola, S.; Moroni, L.; et al. Thyroid-on-a-Chip: An Organoid Platform for In Vitro Assessment of Endocrine Disruption. Adv. Healthc. Mater. 2022, 2201555. [Google Scholar] [CrossRef]
- Zhang, F.; Qu, K.Y.; Zhou, B.; Luo, Y.; Zhu, Z.; Pan, D.J.; Cui, C.; Zhu, Y.; Chen, M.L.; Huang, N.P. Design and fabrication of an integrated heart-on-a-chip platform for construction of cardiac tissue from human iPSC-derived cardiomyocytes and in situ evaluation of physiological function. Biosens. Bioelectron. 2021, 179, 113080. [Google Scholar] [CrossRef]
- Lee, K.K.; McCauley, H.A.; Broda, T.R.; Kofron, M.J.; Wells, J.M.; Hong, C.I. Human stomach-on-a-chip with luminal flow and peristaltic-like motility. Lab Chip 2018, 18, 3079–3085. [Google Scholar] [CrossRef] [PubMed]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.B.; Whisler, J.A.; Fröse, J.; Yu, C.; Shin, Y.; Kamm, R.D. On-chip human microvasculature assay for visualization and quantification of tumor cell extravasation dynamics. Nat. Protoc. 2017, 12, 865–880. [Google Scholar] [CrossRef]
- Dellaquila, A.; Thomée, E.K.; McMillan, A.H.; Lesher-Pérez, S.C. Chapter 4—Lung-on-a-chip platforms for modeling disease pathogenesis. In Organ-on-a-Chip; Hoeng, J., Bovard, D., Peitsch, M.C., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 133–180. [Google Scholar]
- Banaeiyan, A.A.; Theobald, J.; Paukštyte, J.; Wölfl, S.; Adiels, C.B.; Goksör, M. Design and fabrication of a scalable liver-lobule-on-a-chip microphysiological platform. Biofabrication 2017, 9, 015014. [Google Scholar] [CrossRef] [PubMed]
- Ashammakhi, N.; Wesseling-Perry, K.; Hasan, A.; Elkhammas, E.; Zhang, Y.S. Kidney-on-a-chip: Untapped opportunities. Kidney Int. 2018, 94, 1073–1086. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.N.; J. Green, B.; M. Altamentova, S.; V. Rocheleau, J. A microfluidic device designed to induce media flow throughout pancreatic islets while limiting shear-induced damage. Lab Chip 2013, 13, 4374–4384. [Google Scholar] [CrossRef]
- Sharma, K.; Dhar, N.; Thacker, V.V.; Simonet, T.M.; Signorino-Gelo, F.; Knott, G.W.; McKinney, J.D. Dynamic persistence of UPEC intracellular bacterial communities in a human bladder-chip model of urinary tract infection. eLife 2021, 10, e66481. [Google Scholar] [CrossRef]
- Polini, A.; Moroni, L. The convergence of high-tech emerging technologies into the next stage of organ-on-a-chips. Biomater. Biosyst. 2021, 1, 100012. [Google Scholar] [CrossRef] [PubMed]
- De Chiara, F.; Ferret-Miñana, A.; Ramón-Azcón, J. The Synergy between Organ-on-a-Chip and Artificial Intelligence for the Study of NAFLD: From Basic Science to Clinical Research. Biomedicines 2021, 9, 248. [Google Scholar] [CrossRef] [PubMed]
- Petreus, T.; Cadogan, E.; Hughes, G.; Smith, A.; Pilla Reddy, V.; Lau, A.; O’Connor, M.J.; Critchlow, S.; Ashford, M.; Oplustil O’Connor, L. Tumour-on-chip microfluidic platform for assessment of drug pharmacokinetics and treatment response. Commun. Biol. 2021, 4, 1–11. [Google Scholar] [CrossRef]
- Li, J.; Chen, J.; Bai, H.; Wang, H.; Hao, S.; Ding, Y.; Peng, B.; Zhang, J.; Li, L.; Huang, W. An Overview of Organs-on-Chips Based on Deep Learning. Research 2022, 2022, 9869518. [Google Scholar] [CrossRef] [PubMed]
- Paek, K.; Kim, S.; Tak, S.; Kim, M.K.; Park, J.; Chung, S.; Park, T.H.; Kim, J.A. A high-throughput biomimetic bone-on-a-chip platform with artificial intelligence-assisted image analysis for osteoporosis drug testing. Bioeng. Transl. Med. 2023, 8, e10313. [Google Scholar] [CrossRef] [PubMed]
- Oliver, C.R.; Altemus, M.A.; Westerhof, T.M.; Cheriyan, H.; Cheng, X.; Dziubinski, M.; Wu, Z.; Yates, J.; Morikawa, A.; Heth, J.; et al. A platform for artificial intelligence based identification of the extravasation potential of cancer cells into the brain metastatic niche. Lab Chip 2019, 19, 1162–1173. [Google Scholar] [CrossRef]
- Oliver, C.R.; Westerhof, T.M.; Castro, M.G.; Merajver, S.D. Quantifying the Brain Metastatic Tumor Micro-Environment using an Organ-On-A Chip 3D Model, Machine Learning, and Confocal Tomography. J. Vis. Exp. JoVE 2020, 162, e61654. [Google Scholar] [CrossRef]
- Chong, L.H.; Ching, T.; Farm, H.J.; Grenci, G.; Chiam, K.H.; Toh, Y.C. Integration of a microfluidic multicellular coculture array with machine learning analysis to predict adverse cutaneous drug reactions. Lab Chip 2022, 22, 1890–1904. [Google Scholar] [CrossRef]
- Jena, B.P.; Gatti, D.L.; Arslanturk, S.; Pernal, S.; Taatjes, D.J. Human skeletal muscle cell atlas: Unraveling cellular secrets utilizing ‘muscle-on-a-chip’, differential expansion microscopy, mass spectrometry, nanothermometry and machine learning. Micron 2019, 117, 55–59. [Google Scholar] [CrossRef]
- Shannon, M.J.; Mace, E.M. Natural Killer Cell Integrins and Their Functions in Tissue Residency. Front. Immunol. 2021, 12, 647358. [Google Scholar] [CrossRef] [PubMed]
- Parlato, S.; De Ninno, A.; Molfetta, R.; Toschi, E.; Salerno, D.; Mencattini, A.; Romagnoli, G.; Fragale, A.; Roccazzello, L.; Buoncervello, M.; et al. 3D Microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells. Sci. Rep. 2017, 7, 1093. [Google Scholar] [CrossRef] [PubMed]
- Biselli, E.; Agliari, E.; Barra, A.; Bertani, F.R.; Gerardino, A.; De Ninno, A.; Mencattini, A.; Di Giuseppe, D.; Mattei, F.; Schiavoni, G.; et al. Organs on chip approach: A tool to evaluate cancer -immune cells interactions. Sci. Rep. 2017, 7, 12737. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; De Ninno, A.; Mencattini, A.; Mermet-Meillon, F.; Fornabaio, G.; Evans, S.S.; Cossutta, M.; Khira, Y.; Han, W.; Sirven, P.; et al. Dissecting Effects of Anti-cancer Drugs and Cancer-Associated Fibroblasts by On-Chip Reconstitution of Immunocompetent Tumor Microenvironments. Cell Rep. 2018, 25, 3884–3893.e3. [Google Scholar] [CrossRef]
- Comes, M.C.; Casti, P.; Mencattini, A.; Di Giuseppe, D.; Mermet-Meillon, F.; De Ninno, A.; Parrini, M.C.; Businaro, L.; Di Natale, C.; Martinelli, E. The influence of spatial and temporal resolutions on the analysis of cell-cell interaction: A systematic study for time-lapse microscopy applications. Sci. Rep. 2019, 9, 6789. [Google Scholar] [CrossRef] [PubMed]
- Mathur, L.; Ballinger, M.; Utharala, R.; Merten, C.A. Microfluidics as an Enabling Technology for Personalized Cancer Therapy. Small 2020, 16, 1904321. [Google Scholar] [CrossRef] [PubMed]
- Blasiak, A.; Khong, J.; Kee, T. CURATE.AI: Optimizing Personalized Medicine with Artificial Intelligence. SLAS Technol. 2020, 25, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Blasiak, A.; Truong, A.T.L.; Wang, P.; Hooi, L.; Chye, D.H.; Tan, S.B.; You, K.; Remus, A.; Allen, D.M.; Chai, L.Y.A.; et al. IDentif.AI-Omicron: Harnessing an AI-Derived and Disease-Agnostic Platform to Pinpoint Combinatorial Therapies for Clinically Actionable Anti-SARS-CoV-2 Intervention. ACS Nano 2022, 16, 15141–15154. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, K.; Rather, G.M.; Lin, Z.; Sui, J.; Xie, P.; Le, T.; Bertino, J.R.; Javanmard, M. Toward point-of-care assessment of patient response: A portable tool for rapidly assessing cancer drug efficacy using multifrequency impedance cytometry and supervised machine learning. Microsyst. Nanoeng. 2019, 5, 1–11. [Google Scholar] [CrossRef]
- Mencattini, A.; Di Giuseppe, D.; Comes, M.C.; Casti, P.; Corsi, F.; Bertani, F.R.; Ghibelli, L.; Businaro, L.; Di Natale, C.; Parrini, M.C.; et al. Discovering the hidden messages within cell trajectories using a deep learning approach for in vitro evaluation of cancer drug treatments. Sci. Rep. 2020, 10, 7653. [Google Scholar] [CrossRef]
- Owh, C.; Ho, D.; Loh, X.J.; Xue, K. Towards machine learning for hydrogel drug delivery systems. Trends Biotechnol. 2023, 41, 476–479. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, H.-F.; Podder, S.; Chen, P.-Y. Microsystem Advances through Integration with Artificial Intelligence. Micromachines 2023, 14, 826. https://doi.org/10.3390/mi14040826
Tsai H-F, Podder S, Chen P-Y. Microsystem Advances through Integration with Artificial Intelligence. Micromachines. 2023; 14(4):826. https://doi.org/10.3390/mi14040826
Chicago/Turabian StyleTsai, Hsieh-Fu, Soumyajit Podder, and Pin-Yuan Chen. 2023. "Microsystem Advances through Integration with Artificial Intelligence" Micromachines 14, no. 4: 826. https://doi.org/10.3390/mi14040826
APA StyleTsai, H.-F., Podder, S., & Chen, P.-Y. (2023). Microsystem Advances through Integration with Artificial Intelligence. Micromachines, 14(4), 826. https://doi.org/10.3390/mi14040826